Ever since the Stone age, man has been swotting and evolving himself inspired by the most singularly crowned source which is Mother Nature itself. All the inventions and discoveries of the decades are somehow influenced by the elegant wonders of nature. But the most wonderful thing that nature has nurtured is Life itself. The complex, the simple, the tiny, and the big all are an extraordinary bioengineering feat of Mother Nature. However, the most stupendous bioengineering feat is the Human Body.
Just one living cell in the human body is, more complex than New York city
Linus Pauling
This Complicated architecture offers endless possibilities for innovation. It possesses a treasure trove of things that can be manufactured by itself only. For centuries we humans have been trying to synthesize these materials in the laboratory. And on the 10th of September, the Breakthrough prize of $3M in Life Sciences (it’s a huge amount!!) was awarded to Dr. David Baker for synthesizing new kinds of protein through a Unique method. These proteins can also have a huge potential in therapeutic cure strategies of various diseases including Covid-19.


Virus and Its Attachments with You
Viruses are a very dangerous and devilish (in my opinion!) class of organisms that exist on earth. They cannot survive and as such need a host to thrive. Now I believe there’s no introduction needed for one of the most potent and lethal viruses of today, and of course famous too is the SARS-CoV-2 virus.
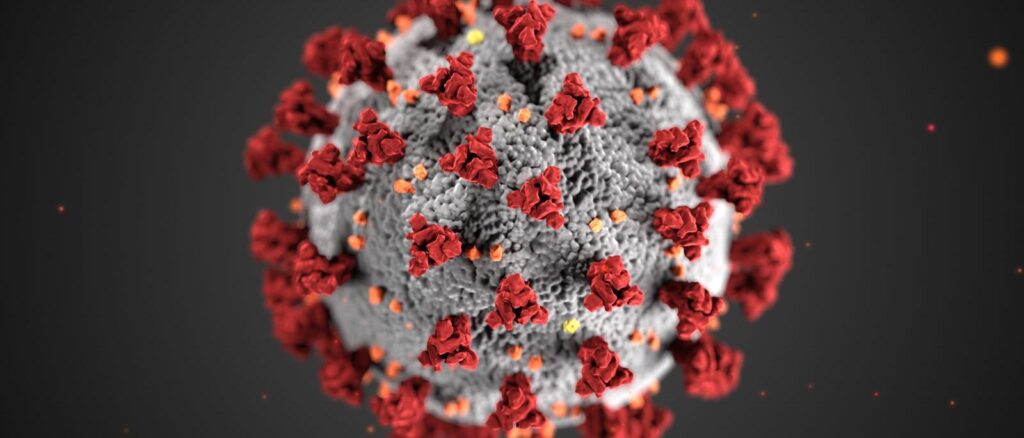
A SARS-CoV-2 virus is dotted all over their surface with Spike Glycoprotein domains or ‘S’ domain giving them a ‘crown’ like appearance giving them their distinctive name- “Corona Viruses”. Each ‘S ’ domain has around 20nm surface projection surrounding the periphery of Coronaviruses. Every ‘S’ domain has a small part on their surface which is essential to interact with endogenous receptors known as “Receptor Binding Domain”. How exactly does the virus bind?
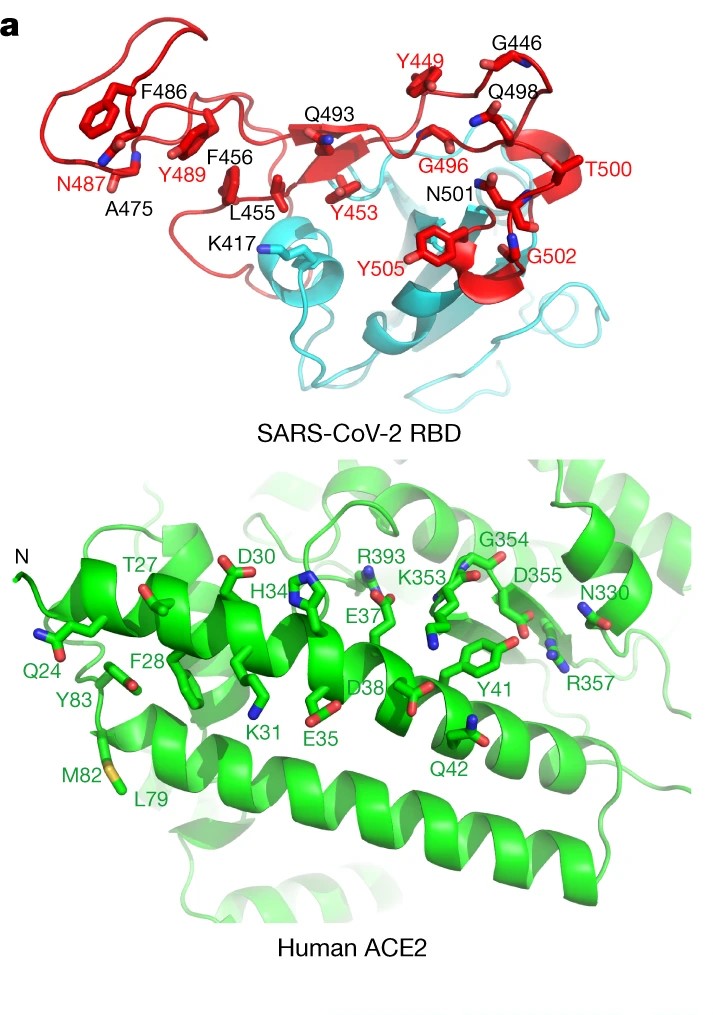
RBD’s help the viruses to dock to the host cell just like rockets that dock to ISS. More importantly, they are the target sites for neutralizing antibodies and other therapeutic strategies. All the treatments and cures, target the RBD of any virus to prevent it from docking to the host cell and minimizing the chances of infection and virus replication.
Now if a person is infected, the virus binds to the host cell and fuses with it. The ribosome processes the RNA of the virus and starts replicating the virus. Once the ribosome creates viral proteins, it cannot be utilized unless it is cleaved by a protein called “Protease”. The work of this protein is to break the viral polypeptide chain into functional proteins. Now the Protease is a dimer of two identical protein chains and binding any one of them to a drug will prevent them from dimerizing, ultimately breaking the chain. This is another therapeutic strategy to prevent infection.
The DE NOVO Approach
The researchers at the Institute for Protein Design, University of Washington, and the Baker Lab accepted the challenge. The team decided to bring a new player in the game, the Classical Supercomputer. Sorry, Quantum Computer, our old friend is here to stay. So the team used special software to design and synthesize proteins using two DE NOVO (to design from scratch) approaches.
[Image- Sciencemag.org]
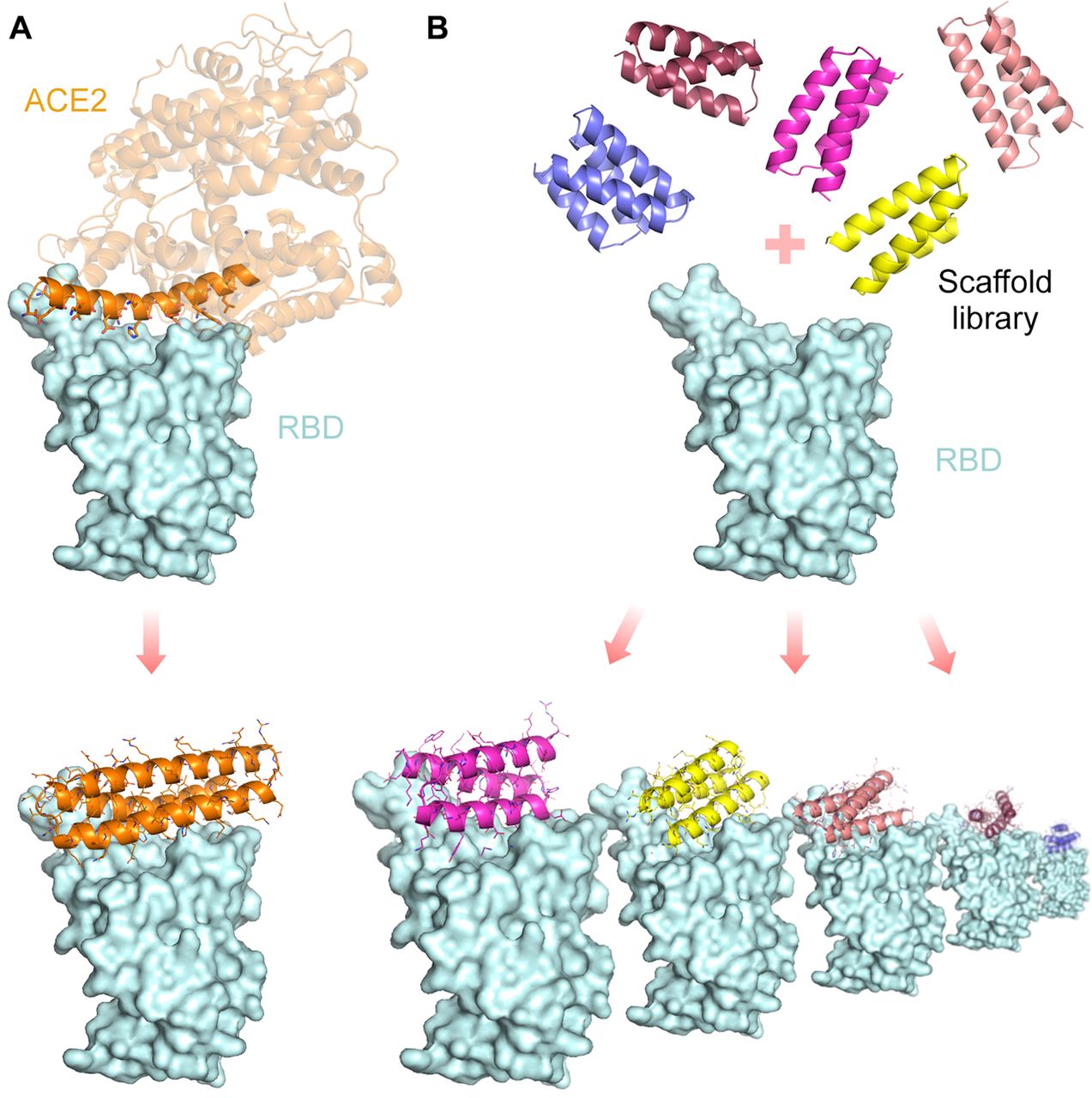
Their main goal was to synthesize proteins that would compete with ACE2 to bind with the RBD of the virus and inhibit its replication. Therefore they decided to make high-affinity protein minibinders. First, they incorporated the alpha helix from ACE2 into small designed proteins that make additional interactions with RBD. For this, they designed scaffolds (biologically temporary structures) around the ACE2 helix to map out its design. Next, they designed proteins from scratch. An advantage of the latter approach is that it offers a range of possibilities and greater diversities for design. For the first approach, the used the Rosetta blueprint binder while for the second approach they used RIF docking and large miniprotein binders.
Armed with a large pool of minibinders, their main aim was to identify the one suitable candidate for the job: ‘Hitman’ (this is what I like to call it! Cool Nah?) The DS method identified three ACE2 helix scaffolded designs and 105 de novo designs.
Improvisation Is Key To Optimization
The modification of ACE2 scaffolded designs by PCR mutagenesis leads to a variant called AHB1 which binds to RBD with an affinity of 1nm. But the design had a low thermostability. Modifying the modified design AHB1, they found one particular design to be thermally stable.
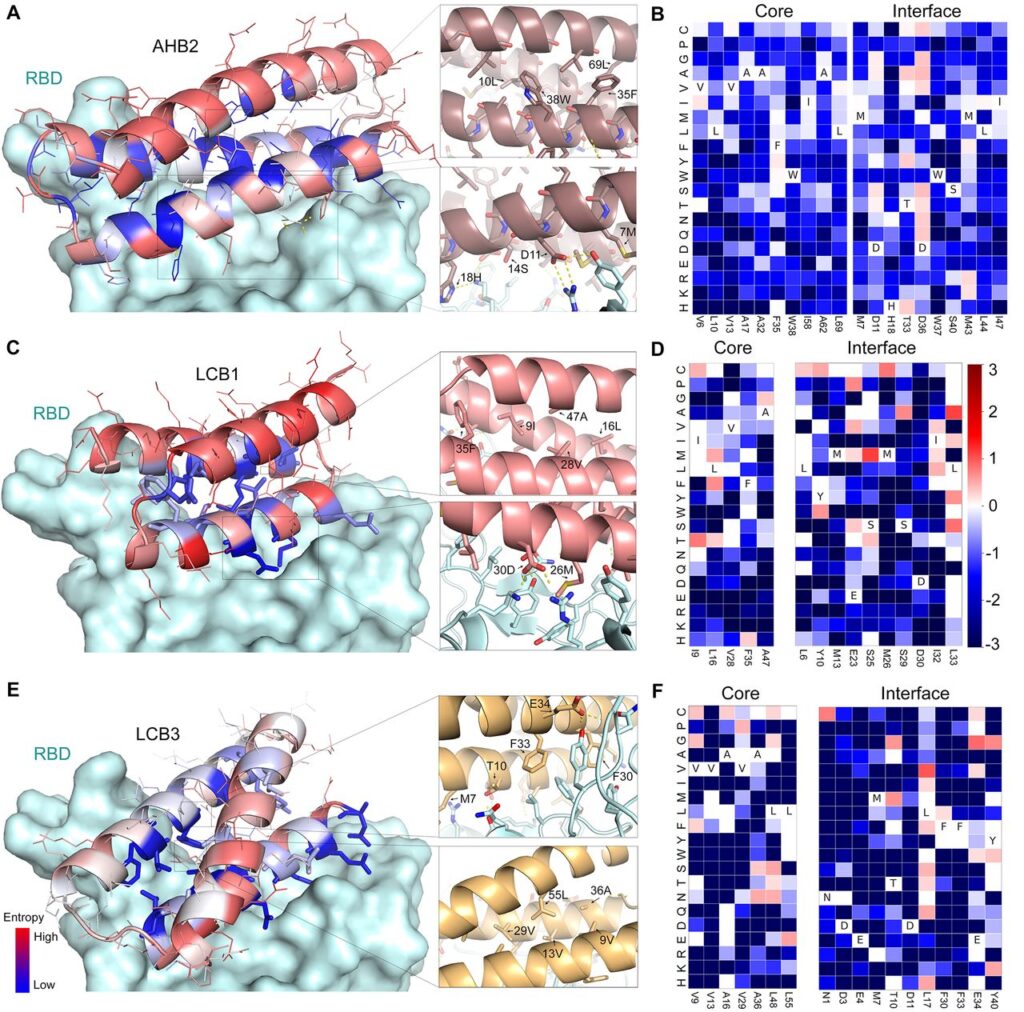
For now, I told you that viruses dock with the ACE2 receptor. Exactly the RBD contains residues(open ends) of amino acid sequence chains that interact with the residues on the ACE2 receptor via hydrogen bonding. Why I’m saying this now will be relevant in the next para.
Out of the 105 minibinders, 50 of them made using approach 2 and 2nd gen ACE2 scaffolded designs were used. They generated SSM libraries in which each of the residues were substituted with 20 of the amino acid one at a time. Using FACS methods, they were able to identify the 8 highest affinity designs. They selected designs: AHB2 and LCB1-LCB8 were found to bind the RBD with high affinity in a manner competed by the ACE2 receptor.
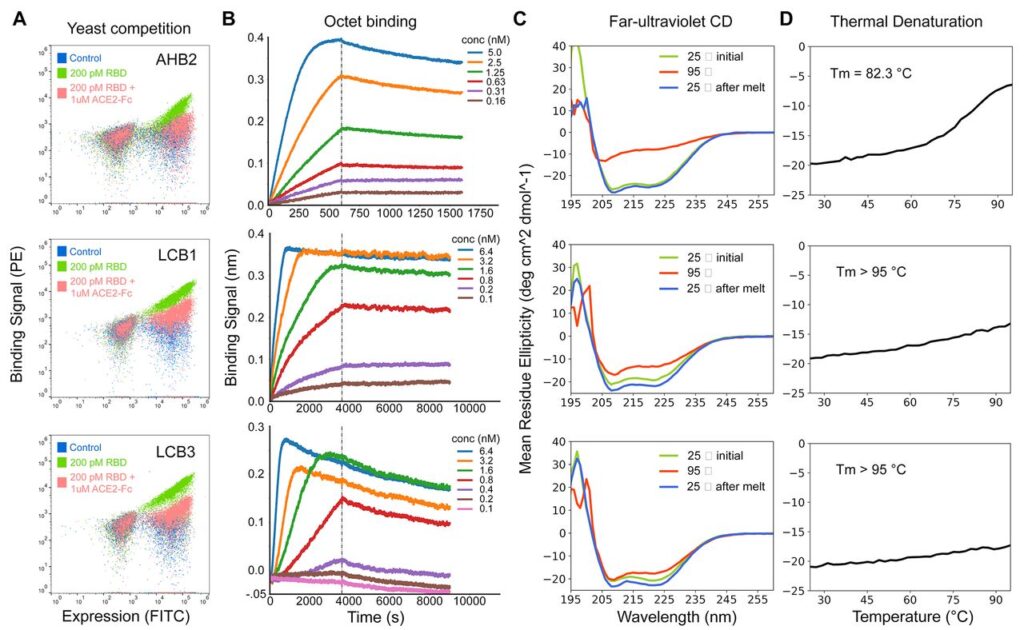
The CD spectra of purified minibinders were compatible with the design versions, and the melting temperature for most was > 90°C. And the best part is that they all retained full binding activity after 14 days at room temperature. AHB1/2 and LCB3 also displayed an affinity towards SARS-CoV RBD. An exciting and detailed analysis of the models by the cryoEM method revealed the consistencies of computational designs with experimental models. This shows that our old friend is not only highly accurate but also at the top of his game!
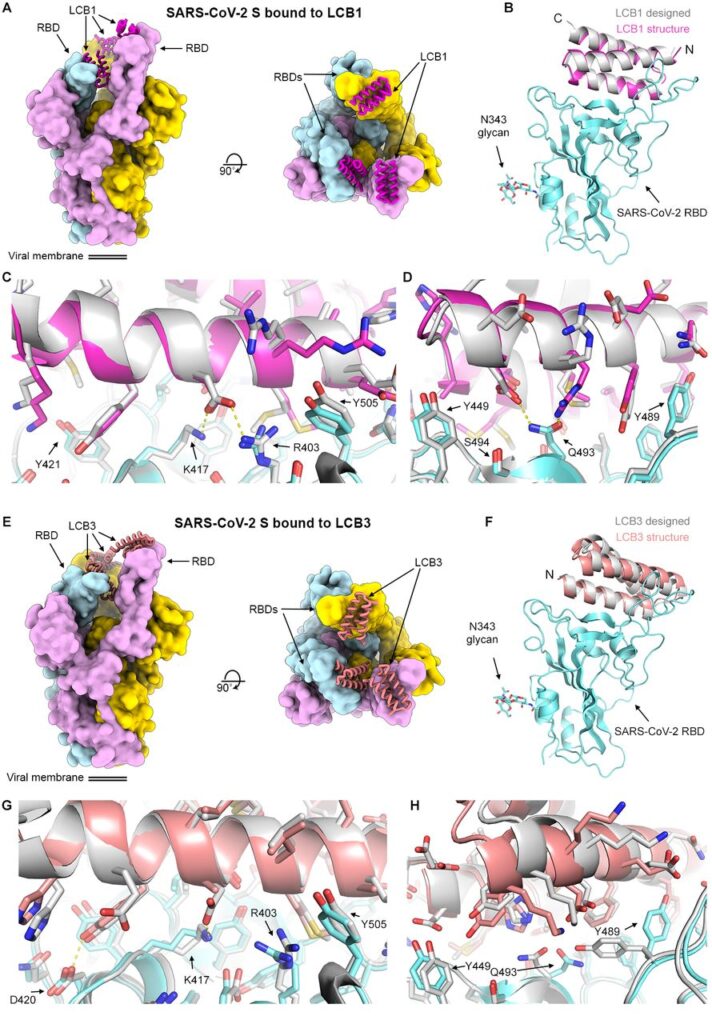
Counterpoising The Menace
Keeping track of all the ‘Hitmans’ and monitoring if they are doing their job properly. They observed that AHB1/AHB2 strongly neutralized the SARS-CoV-2 FFU with IC50 values of 35 nM and 15.5 nM(nano-Molar). On close inspection, the de novo minibinders LHB1, and LHB3 were more potent in nullifying the threat. The observed IC50 values were 23.54 pM and 48.1 pM(pico-Molar). IC50 values refer to half of the max amount of substance needed to inhibit a given biological process or component.
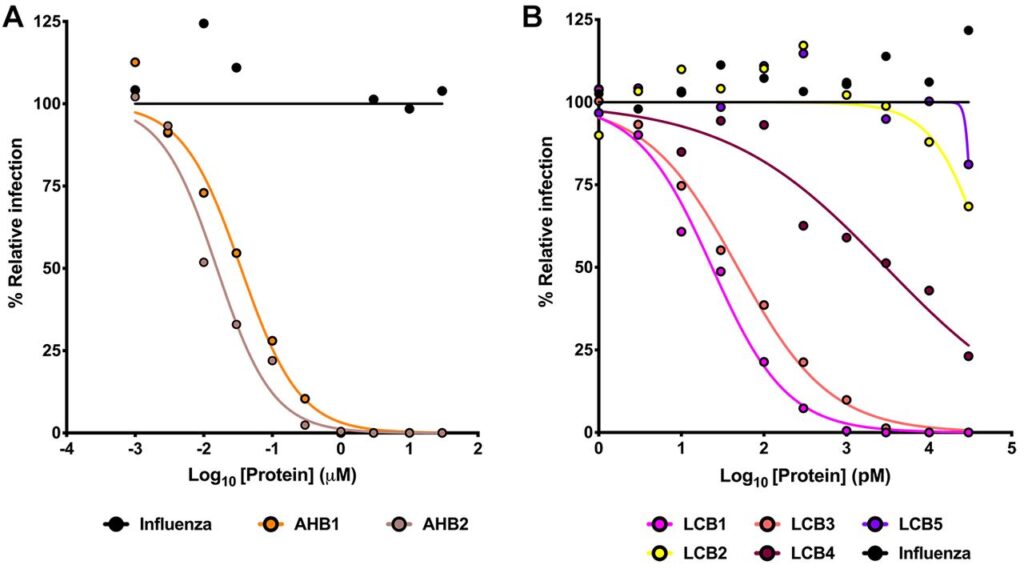
On a mass or molar basis these “Hyperstable Minibinders”, is a term coined by Dr. Baker. He also adds “Owing to the small size and stability of these proteins, they are 20 times more efficient than the most efficient monoclonal antibodies today(used in the same mass)!” The retention of activity even at elevated temperatures indicates that a Temperature controlled chain is not needed.
Binding The Present And Future
The cost of scaling up this technology is very less for much simpler mini proteins than antibodies production. According to Dr. Baker “The small size and stability also make it compliant for direct delivery into the respiratory tract via nebulization or dry aerosol powder.” Here Immunogenicity can be a bit of a problem. Immunogenicity is our body’s response to foreign molecules. So far, they didn’t observe any attack by human cell towards these proteins.
“Timing is very critical witnessing the current situation! Potent therapeutics are needed ASAP!!” He is very excited about his work and has decided to pour in all the prize money in the research. He says “Our lab’s sole aim is to bring today’s Biology out of the Stone Age!” With continuations in the development of the computational field, he believes such advancements would help to prepare against future pandemics.
If You Want Something New, You Have to Stop Doing Something Old!
Peter F. Drucker




[…] Apart from DNA, Proteins are essential biochemical components too. To get more details about “Proteins like never before have been synthesized in the year of 2019”; Click here […]